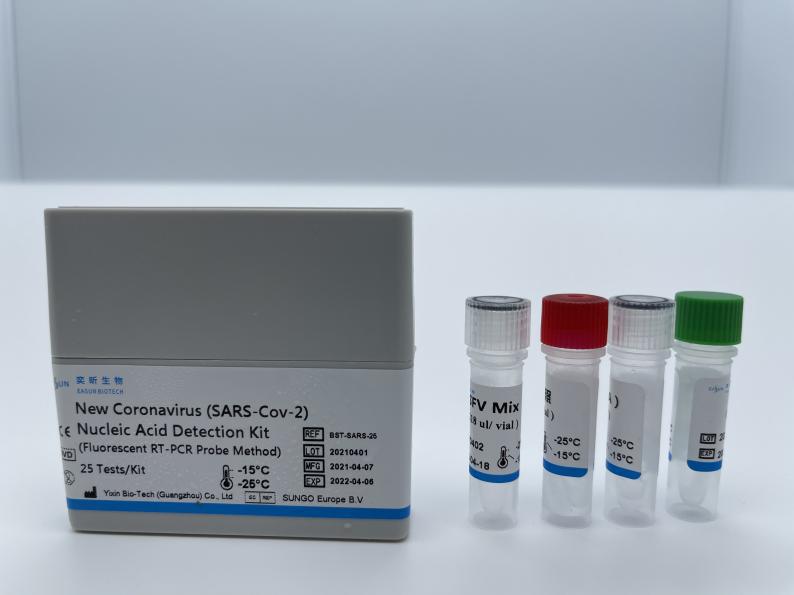
New Coronavirus(SARS-Cov-2) Nucleic Acid Detection Kit
New Coronavirus(SARS-Cov-2) Nucleic Acid Detection Kit
(Fluorescent RT-PCR Probe Method) Product Manual
【Product name 】New Coronavirus(SARS-Cov-2) Nucleic Acid Detection Kit(Fluorescent RT-PCR Probe Method)
【Packaging specifications 】25 Tests/Kit
【Intended usage】
This kit is used for the qualitative detection of nucleic acid from new coronavirus in nasopharyngeal swabs, oropharyngeal (throat) swabs, anterior nasal swabs, mid-turbinate swabs, nasal washes and nasal aspirates from individuals who are suspected of COVID19 by their healthcare provider. The detection of ORF1ab and N genes of the new coronavirus can be used for auxiliary diagnosis and epidemiological monitoring of new coronavirus infection.
【Principles of the procedure 】
This kit is designed for specific TaqMan probes and specific primers designed for the novel coronavirus (SARS-Cov-2) ORF1ab and N gene sequences. The PCR reaction solution contains 3 sets of specific primers and fluorescent probes for specific detection of targets, and an additional set of specific primers and fluorescent probes is used as the internal standard control of the kit for detecting endogenous housekeeping genes.
The test principle is that the specific fluorescent probe is digested and degraded by the exonuclease activity of the Taq enzyme in the PCR reaction, so that the reporter fluorescent group and the quenched fluorescent group are separated, so that the fluorescence monitoring system can receive a fluorescent signal, and then Through the enrichment effect of PCR amplification, the fluorescence signal of the probe reaches a set threshold value-Ct value (Cycle threshold). In the case of no target amplicon, the reporter group of the probe is close to the quenching group. At this time, the fluorescence resonance energy transfer occurs, and the fluorescence of the reporter group is quenched by the quenching group, so that the fluorescent signal cannot be detected by the fluorescent PCR instrument.
In order to monitor the use of reagents during the test, the kit is equipped with positive and negative controls: the positive control contains the target site recombinant plasmid, and the negative control is Distilled water, which is used to monitor environmental pollution. It is recommended to set a positive control and a negative control simultaneously when testing.
【Main components 】
| Cat. No. | BST-SARS-25 | BST-SARS-DR-25 | Components | |
| Name | Specification | Quantity | Quantity | |
| Positive control | 180 μL/vial | 1 | 1 | Artificially constructed plasmids, Distilled water |
| Negative control | 180 μL/vial | 1 | 1 | Distilled water |
| SARS-Cov-2 Mix | 358.5 μL/vial | 1 | / | Specific primer pairs, specific detection fluorescent probes, dNTPs, , MgCl2, KCl, Tris-Hcl, Distilled water, etc |
| Enzyme Mix | 16.5 μL/vial | 1 | / | Taq enzymes, reverse transcriptase,UNG enzymes, etc. |
| SARS-Cov-2 Mix(Lyophilised) | 25 tests/vial | / | 1 | Specific primer pairs, specific detection fluorescent probes, dNTPs,Taq enzymes, reverse transcriptase, Distilled water, etc. |
| 2x Buffer | 375 μL/vial | / | 1 | MgCl2, KCl, Tris-Hcl, Distilled water, etc. |
Note : ( 1) The components in different batch kits cannot be mixed or interchanged.
(2) Prepare your own reagent: Nucleic acid extraction kit.
【Storage conditions and expiration date 】
For BST-SARS-25: Transport and store at -20±5℃ for a long time.
For BST-SARS-DR-25: Transport at room temperature. Store at -20±5℃ for a long time.
Avoid repeated freeze-thaw cycles. The validity period is tentatively set for 12months.
See the label for the date of manufacture and use.
After first opening, the reagent can be stored at -20±5 ° C for no more than 1 month or until the end of the reagent period, whichever date comes first, to avoid repeated freeze-thaw cycles, and the number of reagent freeze-thaw cycles should not exceed 6 times.
【Applicable instrument】 ABI 7500, SLAN-96P, Roche-LightCycler-480.
【Sample requirements 】
1.Applicable sample type: Extracted nucleic acid solution.
2.Sample storage and transportation: Store at-20±5℃for 6 months.Freeze and thaw the samples no more than 6 times.
【Testing method】
1.Nucleic acid extraction
Select a suitable nucleic acid extraction kit to extract viral nucleic acid, and follow the corresponding kit instructions. It is recommended to use a Nucleic acid extraction and purification kit produced by Yixin Bio-Tech (Guangzhou) Co., Ltd. or equivalent nucleic acid purification kit.
2. Reaction reagent preparation
2.1 For BST-SARS-25:
( 1) Remove the SARS-Cov-2 Mix and Enzyme Mix , completely melt at room temperature mix thoroughly by Vortex device and then centrifuge briefly.
(2) 16.5uL Enzyme Mix was added to 358.5uL SARS-Cov-2 Mix and mixed thoroughly to obtain the mixed reaction solution.
(3) Prepare a clean 0.2 mL PCR octal tube and mark it with 15uL of the above mixed reaction solution per well.
(4) Add 15 μL of purified nucleic acid solution, the positive control and the negative control, and carefully cover the octal tube cap.
(5) Mix well by inverting upside down, and quickly centrifuge to concentrate the liquid at the bottom of the tube.
1
2.2 For BST-SARS-DR-25:
( 1) Add 375ul 2x Buffer to SARS-Cov-2 Mix((Lyophilised) to prepare the reaction mix. Mix thoroughly by pipetting and then centrifuge briefly. (Once reaction mix is prepared, it is recommended to store at -20℃ at long-term storage.)
(2) Prepare a clean 0.2 mL PCR octal tube and mark it with 15μL of reaction mix per well.
(3) Add 15μL of purified nucleic acid solution, the positive control and the negative control, and carefully cover the octal tube cap.
(4) Mix well by inverting upside down, and quickly centrifuge to concentrate the liquid at the bottom of the tube.
3. PCR amplification (Please refer to the instrument manual for operation settings.)
3. 1 Place the PCR 8-tube into the sample chamber of the fluorescent PCR instrument, and set the sample to be tested, the positive control and the negative control according to the order of loading.
3.2 Fluorescence detection channel:
( 1) ORF1ab gene selects the detection channel of FAM (Reporter: FAM, Quencher: None).
(2) N gene selects the detection channel of VIC (Reporter: VIC, Quencher: None).
(3) Internal standard gene selects the detection channel of CY5 (Reporter: CY5, Quencher: None).
(4) The Passive Reference is set to ROX.
3.3 PCR program parameter setting:
| Step | Temperature(℃) | Time | Number of cycles | |
| 1 | Reverse transcription reaction | 50 | 15 min | 1 |
| 2 | Taq enzyme activation | 95 | 2.5 min | 1 |
| 3 | Taq enzyme activation | 93 | 10 s | 43 |
| Annealing extension and fluorescence acquisition | 55 | 30 s | ||
After setting, save the file and run the reaction program..
4. Results analysis
After the program ends, the results are automatically saved, and the amplification curve is analyzed. The amplification curve is set to the instrument default threshold.
【Explanation of test results 】
1. Determine the validity of the experiment: The positive control FAM, VIC channel should have a typical amplification curve, and the Ct value is generally less than 34, but may fluctuate due to different threshold settings of different instruments. The negative control FAM, VIC channel should be non-amplified Ct. It is agreed that the above requirements must be met at the same time, otherwise this test is invalid.
2. Result judgment
| FAM/VIC channel | Judgement result |
| Ct<37 | Sample test is positive |
| 37≤Ct<40 | The amplification curve is S-shaped, and suspicious samples need to be re-examined; if the re-examination results are consistent, it is judged as positive, otherwise it is negative |
| Ct≥40 Or No Amplification | Sample test is negative (or below the lower limit of kit detection) |
Note: ( 1) If both the FAM channel and the VIC channel are positive at the same time, the SARS-Cov-2 is determined to be positive.
(2) If the FAM channel or VIC channel is positive and the other channel is negative, the test should be repeated. If it is positive at the same time, it will be judged as SARS-Cov-2 positive, otherwise it will be judged as SARS-Cov-2 negative.